|
|
Load acquired data from .txt file |
| Tags | open☁load☁txt |
A text file is one of the simplest means to store information, being a format outputted by OpenSignals .
In this Jupyter Notebook it will be explained how to load/transpose the data inside .txt file to a Python list, which consists in a step that precedes all processing operations.
1 - Importation of the needed packages
# Package used for loading data from the input text file
from numpy import loadtxt
# biosignalsnotebooks python package
import biosignalsnotebooks as bsnb
2 - Access to electrophysiological signals list
2.1 - Enter biosignalsplux url

2.2 - Navigate through biosignalsplux main page menu and enter in "Signal Samples" page

2.3 - Interactive buttons for accessing each signal sample file
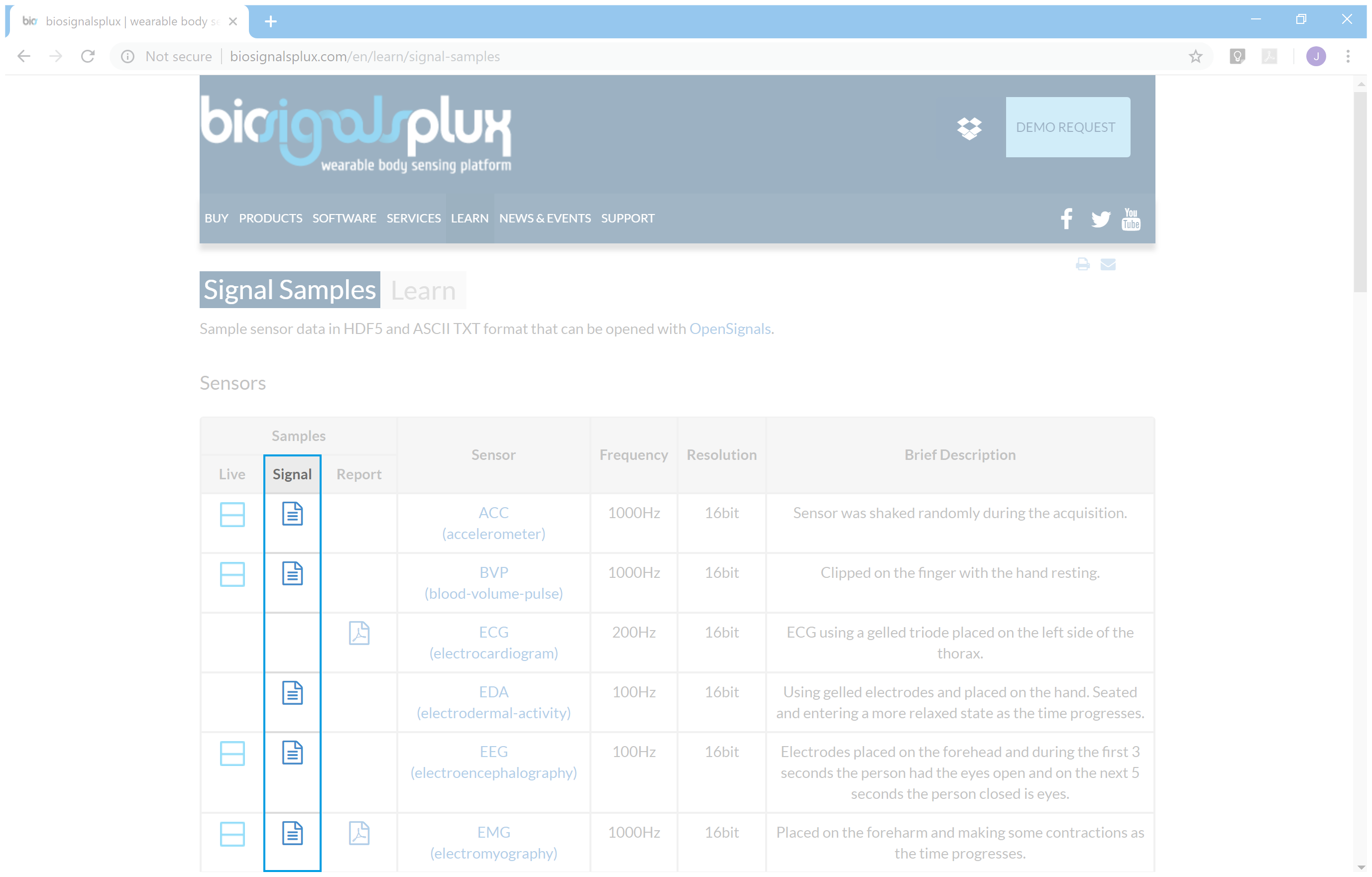
2.4 - File url copy (right-click of the mouse in the desired signal file icon)
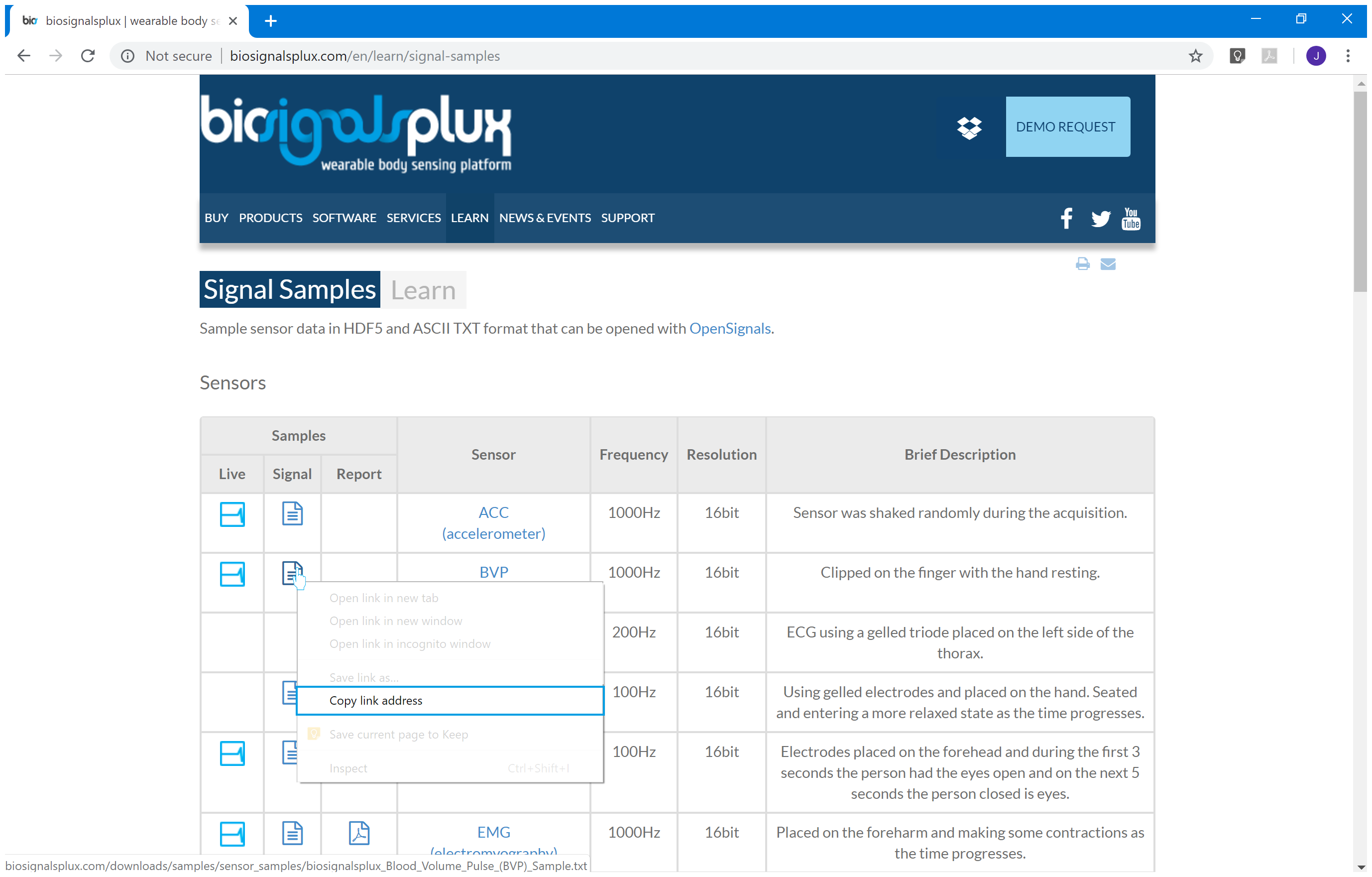
copy_link = 'https://www.biosignalsplux.com/downloads/samples/sensor_samples/biosignalsplux_Electrodermal_Activity_EDA_Sample.txt'
3 - Get file from URL ( http:/downloads/samples/sensor_samples/biosignalsplux_Blood_Volume_Pulse_(BVP)_Sample.txt )
# File download.
bsnb.download(copy_link, out="download_file_name.txt")
4 - Transposition of data to a Python list
data = loadtxt("download_file_name.txt")
5 - Identification of acquisition sampling rate in the file header ("sampling rate" key)

sampling_rate = 1000
6 - Generation of time axis for signal plotting
time = bsnb.generate_time(data, sampling_rate)
7 - Final Output of the loaded data
print (data)
Each line of the list defines a sample acquired at a specific time instant and each column can be the sample number ( nSeq ), digital input ( DI ) or a sample value ( CH1 ), like described in the file header bellow.

The samples of the signal under analysis are stored at the third entry of each list element (index 2).
channel_column = 2
8 - Graphical representation of the signal (raw data)
bsnb.plot(time, data[:, channel_column])
This procedure can be automatically done by load function of biosignalsnotebooks package
Text files are very popular and, like the name suggests, almost all type of contents can be stored here if they can be translated into a text format.
Numpy loadtxt function is very simple and efficient, so, it can be used even for text files not returned by OpenSignals .
We hope that you have enjoyed this guide.
biosignalsnotebooks
is an environment in continuous expansion, so don"t stop your journey and learn more with the remaining
Notebooks
![]() !
!
