|
|
Signal Loading - Working with File Header |
| Tags | load☁metadata☁header |
Each one of the OpenSignals outputted file formats has some metadata linked to it.
Acquisition parameters such as sampling rate, resolution, used channels... can be found in the file metadata. For the .txt file this info is stored in the header while in .h5 files is passed as attributes of the hierarchical objects.
In the current Jupyter Notebook a detailed procedure for accessing file metadata (.txt and .h5) is explained, together with a simplified approach through the use of a biosignalsnotebooks specialized function.
0 - Importation of the needed packages
# biosignalsnotebooks Python package with useful functions that support and complement
# the available Notebooks
import biosignalsnotebooks as bsnb
# Package dedicated to process Python abstract syntax grammar Abstract
# (Abstract Syntax Trees)
from ast import literal_eval
# Package used for accessing .h5 file contents
from h5py import File
1A - Working with a .txt file (generate a dictionary that contains the header info)
1.1A - Open file (creation of a "file" object)
# Specification of the file path
# (in our case is a relative file path but an absolute one can be specified)
relative_file_path = "/signal_samples/bvp_sample.txt"
# Open file
file_txt = open(relative_file_path, "r")
As can be seen, in the previous IFrame and in the following figure, the metadata of our .txt file is placed in line 2.
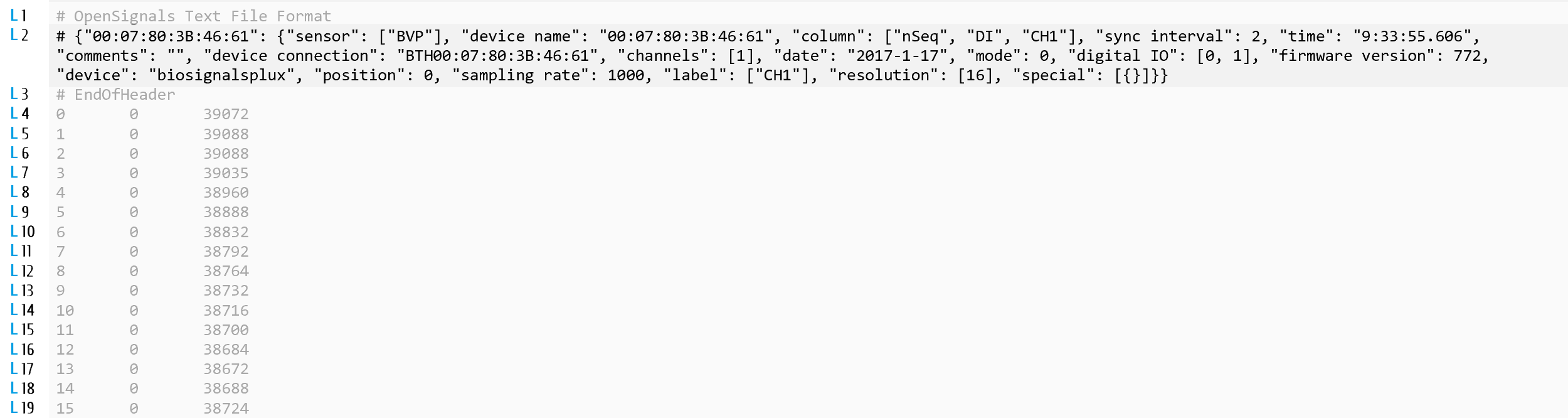
1.2A - Read line 2 of the opened file through "readlines" method of "file_txt" object
# The "readlines" method returns a list where each entry contains a line of the .txt file
txt_data = file_txt.readlines()
# We only need the content of line 2 (entry 1 of "txt_data" list)
metadata = txt_data[1]
# Close file
file_txt.close()
1.3A - Conversion of line content into a dictionary
1.3.1A - Removal of "#" symbol at the beginning of the string, the space between "#" and "{" and the new line command "\n" at the end of the string
Essentially we want to exclude the first two entries of the string (0 and 1) and the last one (-1).metadata_aux = metadata[2:-1]
1.3.2A - Conversion of the remaining content of the original string to a dictionary format (using ast package)
header_txt = literal_eval(metadata_aux)
1B - Working with a .h5 file (generate a dictionary that contains the header info)
1.1B - Load of .h5 file through the creation of a h5py object
# Specification of the file path
# (in our case is a relative file path but an absolute one can be specified)
relative_file_path = "/signal_samples/ecg_20_sec_1000_Hz.h5"
# Creation of h5py object
file_h5 = File(relative_file_path)
1.2B - Determination of the list of available keys (one per device)
available_keys = list(file_h5.keys())
1.3B - Since we are working only with one device, our mac address is stored in the first entry (index 0) of available_keys list
mac = available_keys[0]
1.4B - Access to the first level of file hierarchy
group_lv1 = file_h5.get(mac)
1.5B - Request of the metadata linked to the current hierarchy level
attrs_lv1 = group_lv1.attrs.items()
1.6B - Final conversion into a dictionary
header_h5 = dict(attrs_lv1)
By a simple "key" call it will be possible to access the values highlighted in blue , at topics 1.3.3A and 1.6B.
# Sampling rate of the acquisition contained inside .txt file
sr_txt = header_txt["00:07:80:3B:46:61"]["sampling rate"]
# Sampling rate of the acquisition contained inside .h5 file
sr_h5 = header_h5["sampling rate"]
The previously described procedures, for accessing metadata from .txt and .h5 files, can be easily achieved while loading a file with bsnb.load function, by specifying the input argument called "get_header" as True
# Specification of the file path (in our case is a relative file path but an absolute one can be specified)
relative_file_path = "/signal_samples/ecg_20_sec_1000_Hz.h5"
data, header = bsnb.load(relative_file_path, get_header=True)
As can be understood, for a correct and efficient processing of OpenSignals files it is mandatory to access the information contained inside the header (.txt files) or stored as attributes (.h5 files), like the acquisition sampling rate or device mac-address.
This info (
metadata
![]() ) is always attached to the "real data" as an essential complement.
) is always attached to the "real data" as an essential complement.
We hope that you have enjoyed this guide.
biosignalsnotebooks
is an environment in continuous expansion, so don"t stop your journey and learn more with the remaining
Notebooks
![]() !
!
