|
|
Problems of low sampling rate (aliasing) |
| Tags | record☁sampling rate☁problems |
All data needs to be acquired before the researcher start their processing stage.
The success of the processing stage is deeply dependent on the quality of acquisition. For example, if the chosen
sampling rate
![]() is not adequate for collecting a specific electrophysiological data, we may have an
aliasing problem
is not adequate for collecting a specific electrophysiological data, we may have an
aliasing problem
![]() problem, which makes impossible to extract knowledge.
problem, which makes impossible to extract knowledge.
Accordingly to the
Nyquist Theorem
![]() , to ensure that each signal (EMG, ECG, EDA...) is acquired correctly (avoiding aliasing) the sampling rate should be at least the double of the maximum frequency component present in the signal, being this threshold known as
"Nyquist Rate"
, to ensure that each signal (EMG, ECG, EDA...) is acquired correctly (avoiding aliasing) the sampling rate should be at least the double of the maximum frequency component present in the signal, being this threshold known as
"Nyquist Rate"
![]() .
.
This maximum frequency corresponds to the last power spectrum component (after application of the Fourier Transform) with relevant information.
In the following steps it will be demonstrated how the sampling rate choice affect signal morphology.
0 - The available sampling rates of PLUX acquisition systems lie between 10 and 4000 Hz, accordingly to the firmware version
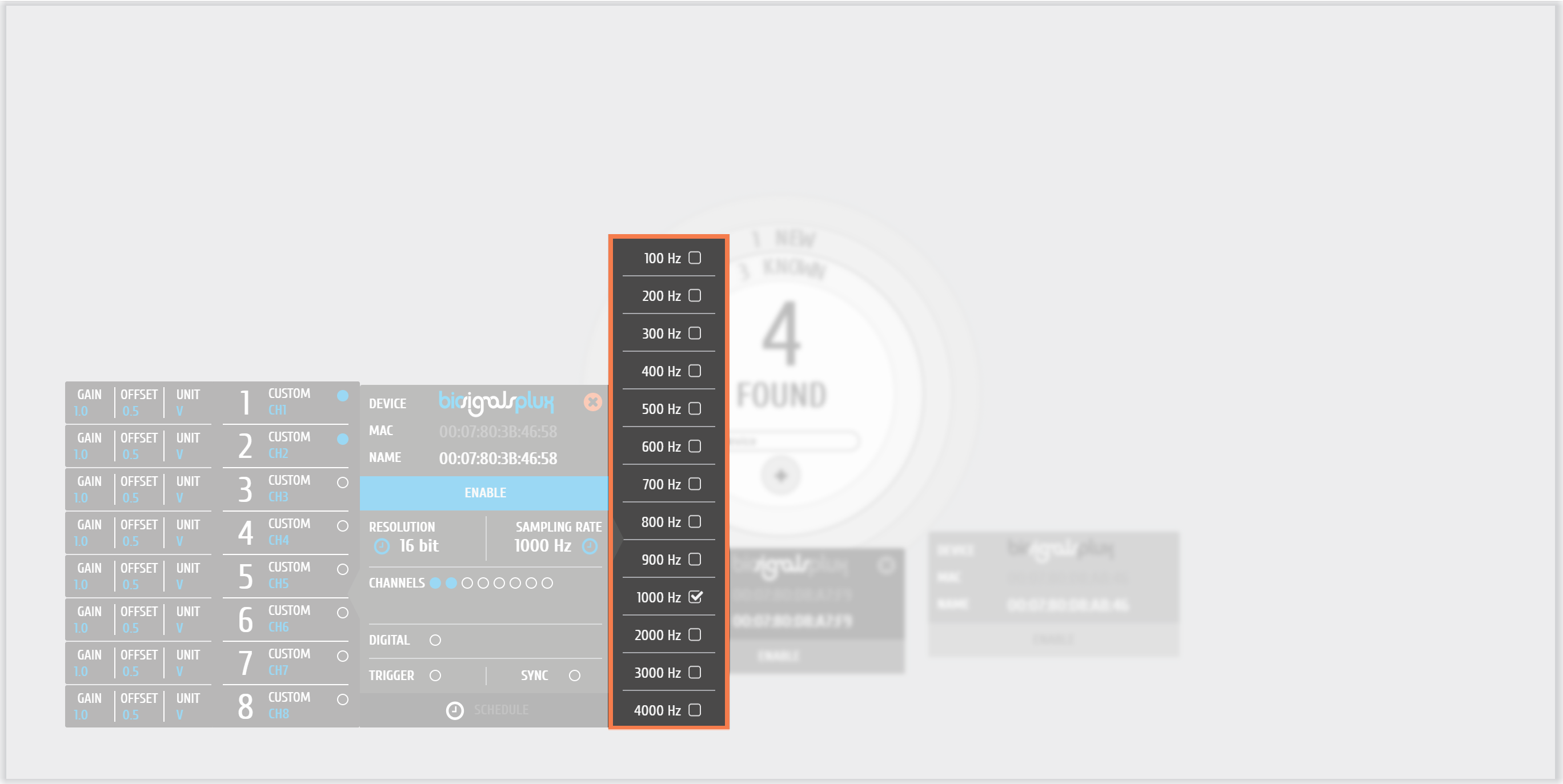
In the following images we can see the effect of sampling rate choice. ECG acquisitions at 10, 100 and 1000 Hz have been done.
1 - Evolution of the ECG acquisition according to the chosen sampling rate ( Available values on PLUX devices )
As demonstrated in the following figure, for this range of sampling rate values, the differences in the signal morphology can be easily observed.- For the acquisition done at 10 Hz, we can't observe ECG complexes or the periodic behavior of cardiac signal;
- With an acquisition at 100 Hz, the aliasing problem was solved and, from that signal, all ECG structures are easily seen;
- Finally with 1000 Hz, the differences in relation to 100 Hz acquisition are more difficult to see, but for example we have a more "impulsive-like" R peak.
For accessing the code instructions responsible for generating the previous figure,
biosignalsnotebooks
users can navigate through the invoked function by clicking on the "Download" button located in Notebook header or by exploring the Python implementation on our
Python package
![]()
Each electrophysiological signal has a characteristic Nyquist Rate, based on the highest informational component (the upper limit of signals pass-band).
These reference values may be found in
Sensors Page
![]() and in the particular case of ECG the bandwidth is presented in
ECG Sensor Page
and in the particular case of ECG the bandwidth is presented in
ECG Sensor Page
![]() (as demonstrated in the following screenshot).
(as demonstrated in the following screenshot).
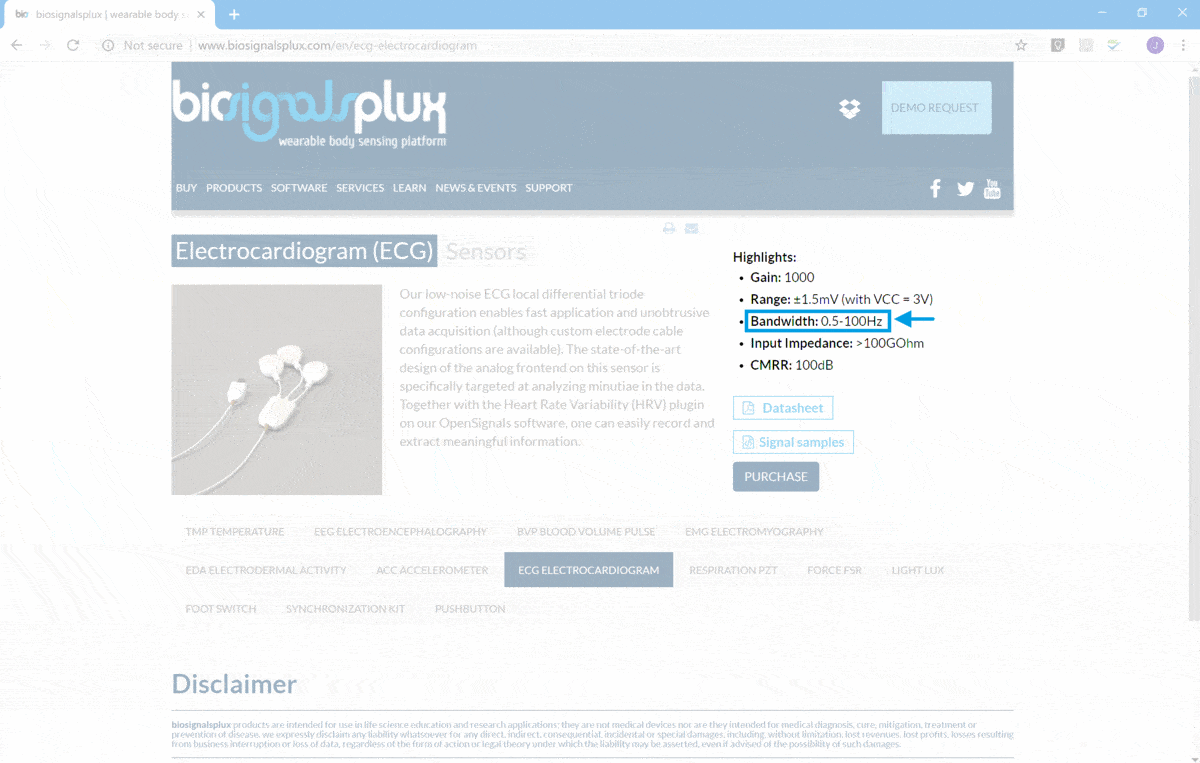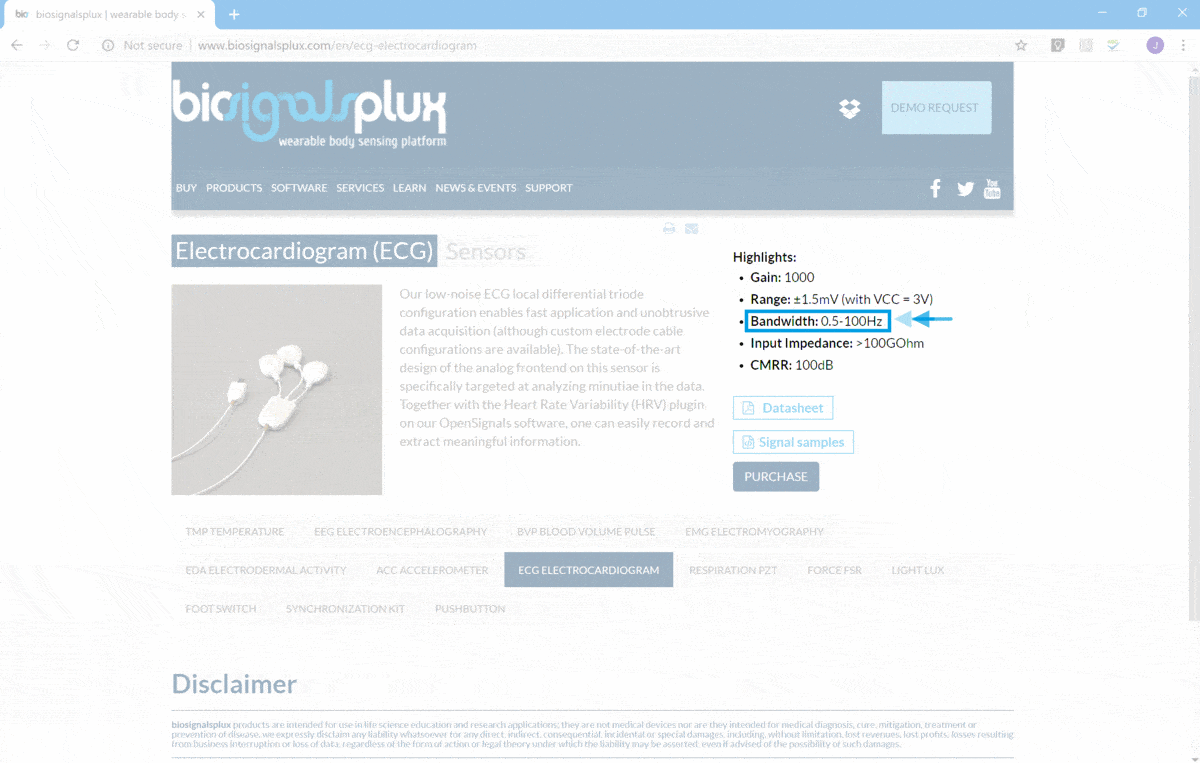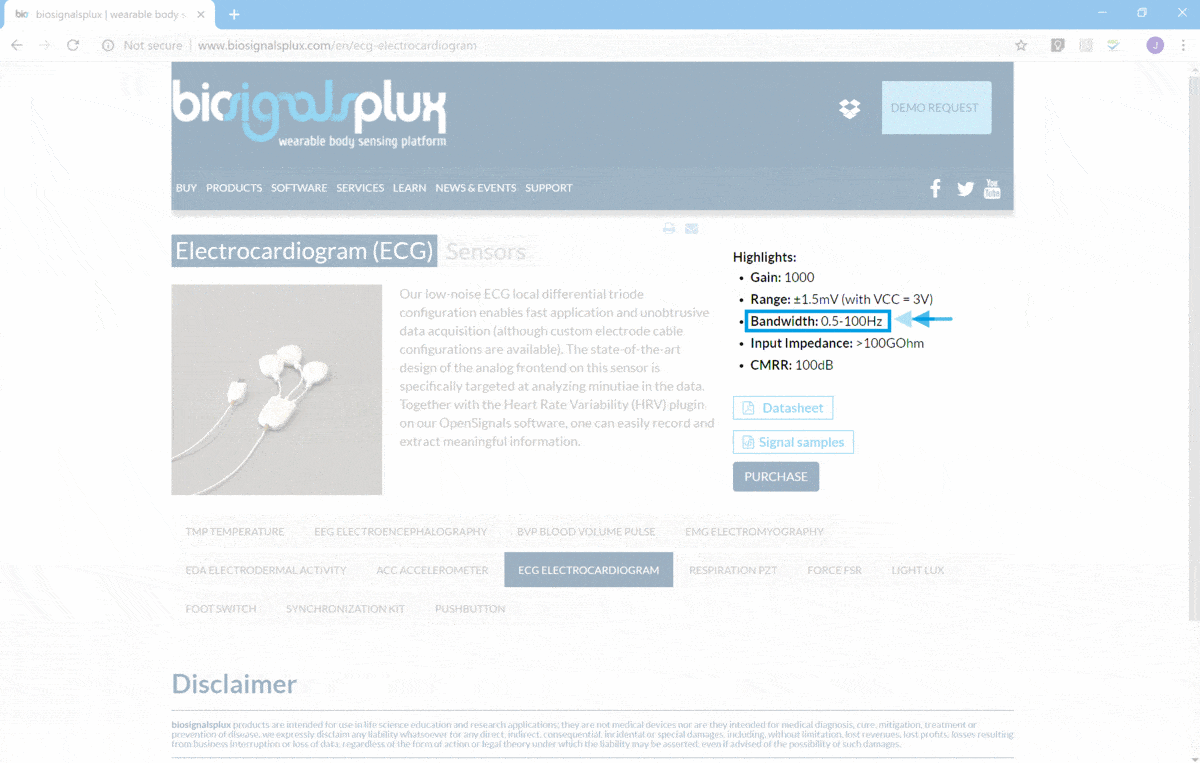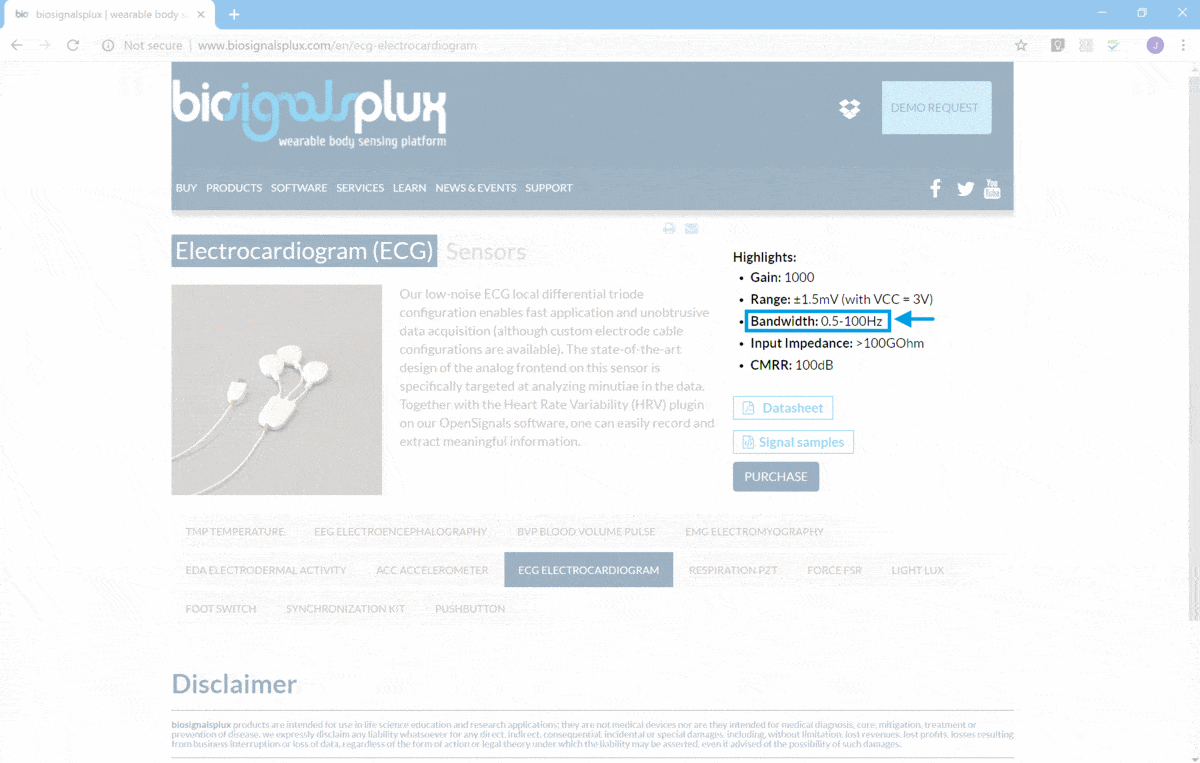
ECG Nyquist Rate will be near 200 Hz, taking into consideration the upper limit of the signal bandwidth (100 Hz).
Sampling rate is one of the most relevant parameters on signal processing research and without this information it is almost impossible to make temporal considerations about the acquired data.
But, as we saw, the researcher should take sampling rate into account not only during the processing, but also before the acquisition !
We hope that you have enjoyed this guide.
biosignalsnotebooks
is an environment in continuous expansion, so don"t stop your journey and learn more with the remaining
Notebooks
![]() !
!
